Introduction
As the name itself says Python Record Linkage Toolkit is used to link the records in the same file or between different data sources. It provides numbers of tool/functions to help in record linkage and deduplication process.
Deduplication
Deduplication is the process of eliminating or removing the redundant data from the given data.
Record Linkage
Record linkage is the process where the data from one source is joined with data from another source that describes the same entity. For example, we can link/join the record Narendra Modi from file_1 with Narendra Damodardas Modi from file_2 as both are referring to the same entity.
Consider this scenario — you are getting two files from two different sources that contain information about the same entity. Let’s say your requirement is to use both the files and generate a report out of both the files using common/similar columns. How do you combine or link records from both files? The recordlinkage toolkit comes to your rescue.
The functionalities provided by recordlinkage library can be broadly classified into five categories. In this blog, we will focus only on Pre-processing, Indexing, and Comparing. I will try to cover Classification and Evaluation in the future article.
- Pre-processing
- Indexing
- Comparing
- Classification
- Evaluation
Once you go through the example below, you will get a good understanding of when Record Linkage Tool Kit can be used.
Installation
pip install recordlinkage
1. Pre-processing
As the name itself says, this pre-processing functionality is used if there is a need to clean the text such as removing whitespace, invalid characters, standardizing the text, etc. You can find the sample code to use pre-processing below. Refer here for more advanced usage on the pre-processing utility.
import pandas as pd
from recordlinkage.preprocessing import clean
names = ['Towards AI',
'Artificial %%Intelligence',
'Chetan (Ambi)',
'$&& Machine %^&* Learning'
]
s = pd.Series(names)
print(clean(s))
0 towards ai
1 artificial intelligence
2 chetan
3 machine learning
2. Indexing
The first line of the code recordlinkage.Index() is a class that will be used to create record pairs based on the different algorithms. Currently, three algorithms are incorporated — full, block, and sortedneighbourhood.
Full
In the below example we are using the full index algorithm for indexing. Then indexer.index() method will create all possible record pairs based on the algorithm chosen. Since we have used full index, it will create n x m possible candidates that can be used in the next steps.
indexer = recordlinkage.Index()
indexer.full()
candidates = indexer.index(dfA, dfB)
print("candidate:", len(candidates))
candidate: 25000000
Blocking
In this method, we choose one or more columns as indices for the comparison. This method is called blocking. Since we are agreeing on one or more columns, the number of possible comparisons will be much lower reducing the computation time. In the below example, we are using given_name column as a blocking variable. As you can see the number of possible candidate links for comparison is reduced significantly.
indexer = recordlinkage.Index()
indexer.block('given_name')
candidate_links = indexer.index(dfA, dfB)
print("candidate_links:", len(candidate_links))
candidate_links: 77249
3. Comparing
Once we used indexing, the next step is to define how you want to compare the columns from both the files. The Recordlinkage compare() method provides advanced usage of how you would like to compare numeric, string, date & geo field types. For example, you may want to compare the ‘given_name’ column from both the files as an exact match, the ‘address’ column as at least 85% match, etc. Finally, the compute() method will compute the similarity and the results are stored in the features.
compare_cl = recordlinkage.Compare()
compare_cl.exact('given_name', 'given_name', label='given_name')
compare_cl.string('surname', 'surname', method='jarowinkler', threshold=0.85, label='surname')
compare_cl.exact('date_of_birth', 'date_of_birth', label='date_of_birth')
compare_cl.exact('suburb', 'suburb', label='suburb')
compare_cl.exact('state', 'state', label='state')
compare_cl.string('address_1', 'address_1', threshold=0.85, label='address_1')
features = compare_cl.compute(candidate_links, dfA, dfB)
- exact compares the record pairs exactly. The similarity is 1 if an exact match otherwise 0.
- string computes the similarity between the strings. It uses different algorithms from the list: jaro, jarowinkler, levenshtein, damerau_levenshtein, qgram or cosine. The default is levenshtein.
- numeric is used to compute the similarity between numeric values. Different algorithms can be used from the list: step, linear, exp, gauss or squared. The default is linear.
Next, we need to find out which records belong to the same entity (matching process). Below is the output after the comparison.
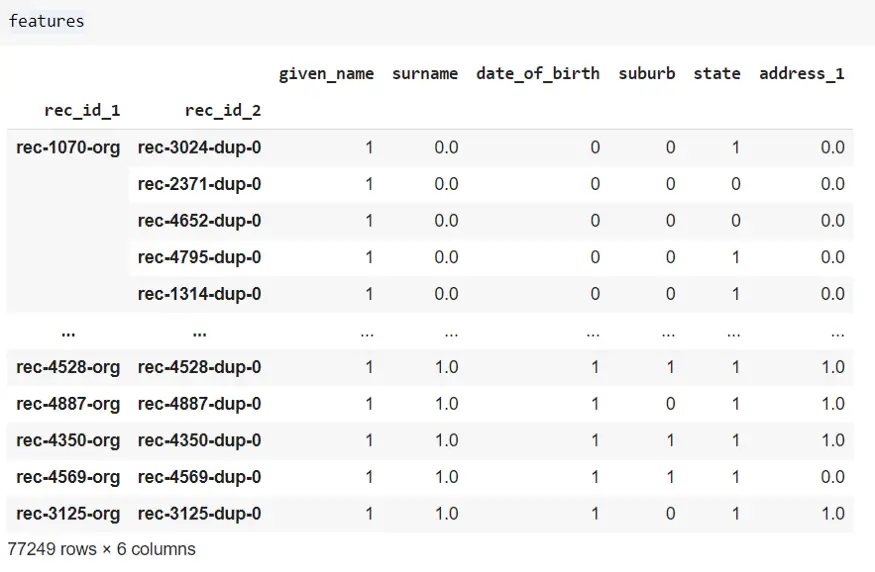
With simple logic, we can filter out similar and non-similar records as below. There are 6 columns/features so one option to filter matched records is for each row to take the sum and filter out based on the sum > 3 (this means at least 4 columns are matched). Similarly, sum ≤3 will be non-similar records.
Below codes says there are 1566 records where all 6 columns are matched/similar, 1332 similar records, and so on between dfA and dfB. And there is a total of 2898 records where at least 4 columns are matched.
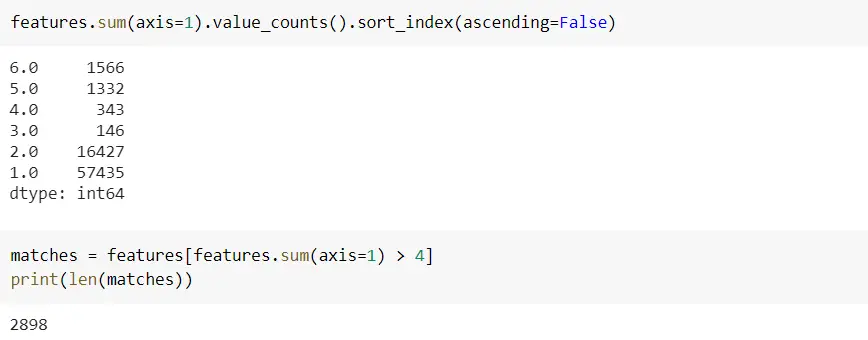
This dataframe shows which record from dfA is matching with the record from dfB.
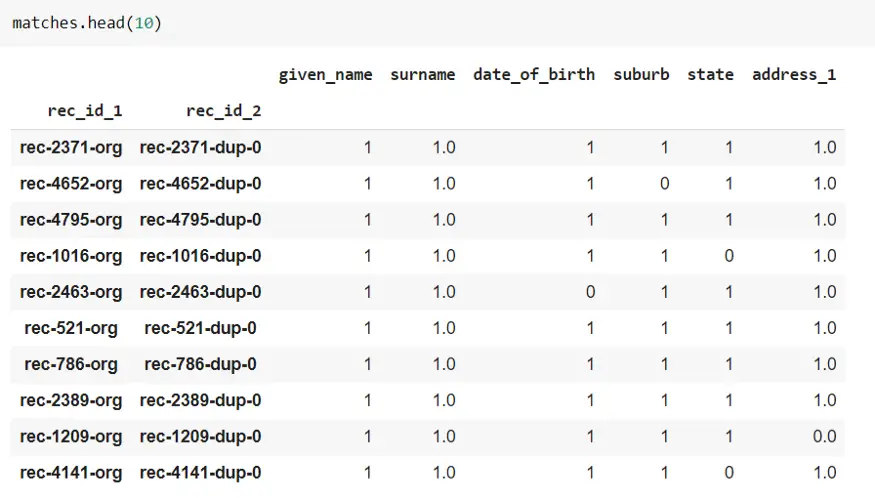
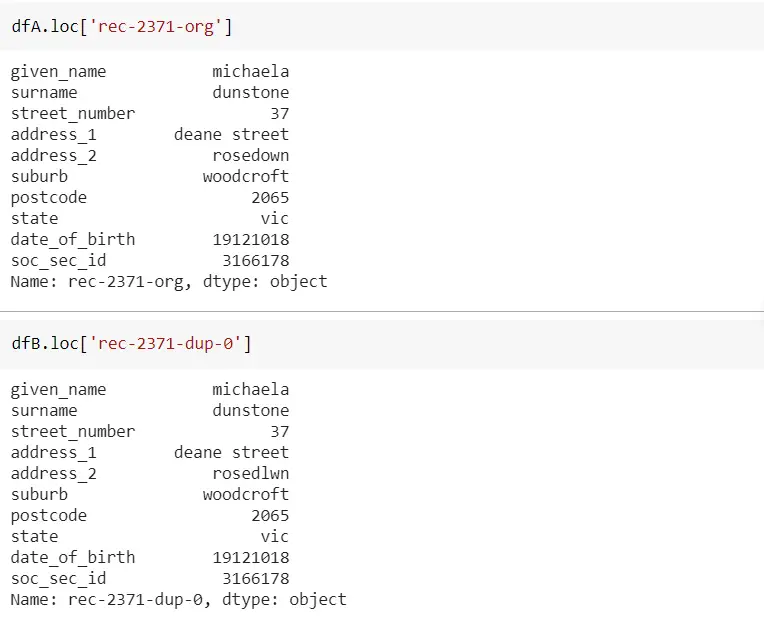
The below screenshot shows individual records from both dfA and dfB for the first matching record from the above figure. As you can see for this matching record, all six columns are matched.
Once we have similar and non-similar records, we can implement the business logic to handle these records to generate the report, etc.
You can refer to the complete code from Github Gist here.
Conclusion
Recordlinkage is the best open-source library I found for record linking and deduplication. In this article, we worked on the example of record linking. If you are looking for deduplication on a single file go through this link and note that it follows almost the same process as record linking.

